From the Bench: Creative Approaches in Cancer Research, Fall 2023
For our readers who enjoy learning about the newest tools and strategies in cancer research, the second installment of our series, “From the Bench,” is here. (Check out the inaugural post here.)
This quarter, we bring you engineered bacteria that detect cancer or tag cancer cells for recognition by immune cells, a strategy to rewire cancer driver genes to induce cell death, implantable microscopic devices to assess drug response, a method to map protein trafficking in living cells, and a way to correlate a cell’s transcriptional profile with its location within a tissue.
Microbes on a Mission: Using Bacteria to Detect and Treat Cancer
Not all bacteria are bad. They may even help us treat cancer, as summarized in a recent blog post. This quarter, we learned about two more efforts to employ bacteria against cancer. In the first study, published in Science, researchers engineered bacteria to sense mutated tumor DNA in their environment.
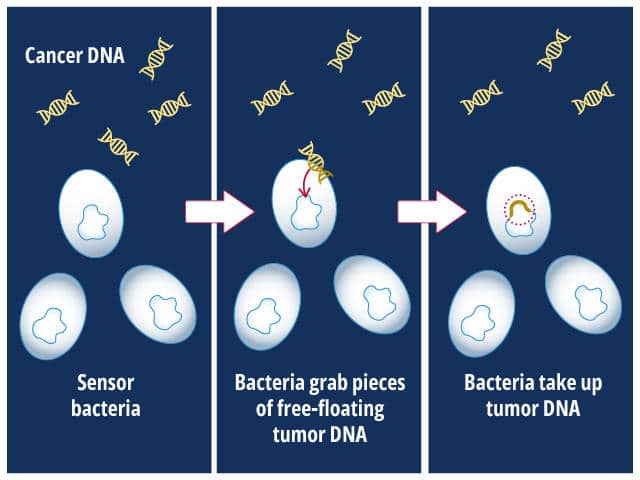
Their design relied on the intrinsic ability of bacteria to internalize DNA from their surroundings.
The bacteria, Acinetobacter baylyi, were engineered to express a gene that made them resistant to the antibiotic kanamycin only if they internalized KRAS-mutated DNA that was shed by cancer cells. To test their design, the researchers implanted the engineered bacteria into the colons of mice that had KRAS-mutated colorectal cancer or into control mice that did not have cancer.
The researchers then isolated bacteria from the mice’s droppings and incubated them in the presence of kanamycin. If the bacteria grew, it meant that KRAS-mutated DNA had been present in the colon and had been taken in by the bacteria. In the proof-of-principle experiment, bacteria from cancer-bearing mice grew in the presence of kanamycin, while the bacteria from control mice did not. The researchers hope that such a strategy could someday be used in the clinic as an early cancer detection tool.
In the second study, also published in Science, researchers used modified Escherichia coli to make solid tumors susceptible to chimeric antigen receptor (CAR) T-cell therapy.
CAR T-cell therapy has been largely unsuccessful against solid tumors due, in part, to the hostile tumor microenvironment that prevents T cells from entering the tumor. To overcome this obstacle, the researchers designed E. coli that would grow exclusively within the immune-privileged and hypoxic microenvironments of solid tumors and release a particular synthetic protein—a protein that corresponding CAR T cells were engineered to recognize.
This two-pronged approach enabled researchers to effectively tag solid tumor cells with a marker that recruited cancer-killing CAR T cells into the difficult-to-reach tumor microenvironment. By using a synthetic peptide to tag the cancer cells, the design also allowed them to avoid using a tumor-specific antigen, making this approach applicable to a variety of tumor types.
The researchers demonstrated that this strategy effectively slowed tumor growth in mouse models of colorectal and breast cancers. However, they noted that further research is needed to understand how safe and effective the design would be in patients.
Attracting Opposites: Turning Cancer Driver Genes Against the Cell
Cancer-driving transcription factors can dramatically change the way tumor cells function by promoting the expression of oncogenes or blocking the expression of tumor suppressor genes. In a recent study in Nature, researchers showed how they can turn these transcription factors into their own worst enemies.
They designed a type of molecule called a transcriptional chemical inducer of proximity (TCIP), which binds a cancer-driving transcription factor to another transcription factor that performs the opposite function. As an example, the researchers selected the driver gene BCL6, which normally suppresses the expression of cell death genes in many human cancers. They employed a TCIP called TCIP1 that joins BCL6 to the transcriptional activator BRD4, forcing BRD4 to activate genes that are typically silenced by BCL6.
This transcriptional rewiring had a profound effect on the viability of cell models of diffuse large B-cell lymphoma and other hematological cancers; increasing BCL6 expression correlated with increased cell death. In addition to stimulating cell death genes, TCIP1 also repressed expression of the oncogene MYC. When administered to healthy mice, TCIP1 showed minimal toxicities. The authors suggested that this system could be applied to other cancer drivers and adapted to nondriver applications, such as overcoming senescence and derepressing potential neoantigen genes.
Lab in a Patient: Multi-drug Screening During Brain Tumor Surgery
For some aggressive tumor types, such as gliomas (a type of brain tumor), it can be challenging to determine which therapy has the best chance of treating each patient’s unique cancer. The applicability of ex vivo techniques, in which a patient’s tumor tissue is screened for drug sensitivity in a laboratory, is limited by technical hurdles and the amount of time necessary to perform such screening.
A recent study in Science Translational Medicine brought the drug screen directly to the tumors instead. While six patients underwent surgery to remove their gliomas, researchers implanted tiny chambers containing concentrated glioma drugs into their tumors. The chambers (approximately 1 inch by 0.7 mm, the width of mechanical pencil lead) were placed at the start of surgery. While surgeons removed other parts of the tumor over an average of 2.3 hours, the drugs diffused from the chambers into nearby tumor tissue, creating a dose-response gradient. The surgeons removed the chamber-containing tumor pieces last, and researchers analyzed the tissue for evidence of cell death and DNA damage.
Implantation of the chambers did not cause any side effects and extended the surgeries by an average of 32.1 minutes. Evidence of drug efficacy correlated well with observed clinical responses; for example, some patients whose drug screens predicted sensitivity to standard-of-care temozolomide experienced clinical responses to temozolomide, and those whose drug screen results suggested temozolomide resistance had either stopped responding to temozolomide prior to surgery or did not respond to adjuvant temozolomide.
While more research is needed to understand the wider implications, the authors emphasized that the one glioma that had developed temozolomide resistance before surgery did not show vulnerability to other tested drugs. This matches previous observations of clinical multidrug resistance, they said, suggesting the critical importance of selecting an effective first-line treatment.
From Here to There: Tracking Protein Movement Within a Cell
Proteins traffic between different cellular compartments and even between different cells to carry out their multitude of functions. The unbiased identification of proteins that shuttle between distinct sites of interest could shed light on the proteins involved in various cellular processes—including those that promote cancer—and help researchers uncover new drug targets.
Recently, researchers reported in Cell a new method, which they call TransitID (for trafficking analysis by sequential incorporation of tags for identification), to map protein trafficking within and between living cells. The method involves pulsing the origin organelle or cell with one label (TurboID) and after some time, pulsing the destination organelle or cell with a second label (APEX).
Proteins that traveled from the origin to the destination during the experiment will be marked by both labels. Researchers can then isolate the dual-labeled proteins and identify them by mass spectrometry.
To demonstrate the utility of their method, the authors of the study used TransitID to identify proteins that traveled between the cytosol and mitochondria, the cytosol and nucleus, the nucleolus and stress granules, and between macrophages and cancer cells under different conditions.
Location, Location, Location: A New Method to Correlate Transcriptional and Spatial Information at the Single-cell Level
With the advent of single-cell RNA sequencing (scRNA-seq) technology, researchers have gained the ability to zoom in on the transcriptional profile of individual cells, which is crucial in dissecting the complexity and heterogeneity of tumor tissues. Since cells exist in neighborhoods, their identity and function are shaped by their location within the tissue, which influences factors such as nutrient and oxygen abundance and the presence of certain external stimuli. To obtain a comprehensive picture of a cell’s identity, it is therefore essential to include information on its physical position.
The authors of a study published in Cell Systems described a method called Segmentation by Exogenous Perfusion (SEEP) to correlate scRNA-seq data with spatial information as a function of the cells’ accessibility within the tissue.
Cell spheroids were exposed to a cell-permeable dye that became fluorescent once internalized by the cells. Penetration of the dye into the spheroids created a fluorescence gradient that correlated with the distance of each cell from the surface. Afterwards, the spheroids were dissociated, the cells were sorted by fluorescence, and their RNA was sequenced via scRNA-seq. Computational methods allowed researchers to interpret the results and correlate the cells’ transcriptional identity with their location.
The authors applied this method to map the transcriptional profiles of high-grade serous ovarian cancer cells in three 3-D models: spheroids, organoids, and patient-derived xenograft tumors.
Cells located in the middle layers of the spheroids had higher expression of genes related to proliferation, while cells on the surface were characterized by higher expression of genes that control inflammation, the ability to migrate to other sites, and interaction with the immune system. Of note, these features were conserved in spheroid-like bodies isolated from patients’ ascites, which are believed to mediate metastatic dissemination.