How AACR Project GENIE Is Helping Unravel the Mysteries of Cancer
Sometimes even the best detectives could use a little assistance with particularly complex mysteries—and the mysteries of cancer are as complex as they come. So, even if cancer researchers have a team full of Sherlock Holmes, they could still use a Dr. Watson to help put things in perspective—or perhaps a genie will do. Over the past decade, AACR Project GENIE has evolved from a repository of tumor genomic and clinal data to become a tool for training artificial intelligence (AI) and machine learning (ML) models, synthesizing data, and predicting drug responses and patient outcomes. This evolution is helping researchers investigate queries about cancer that have the potential to transform care and improve patient outcomes worldwide.
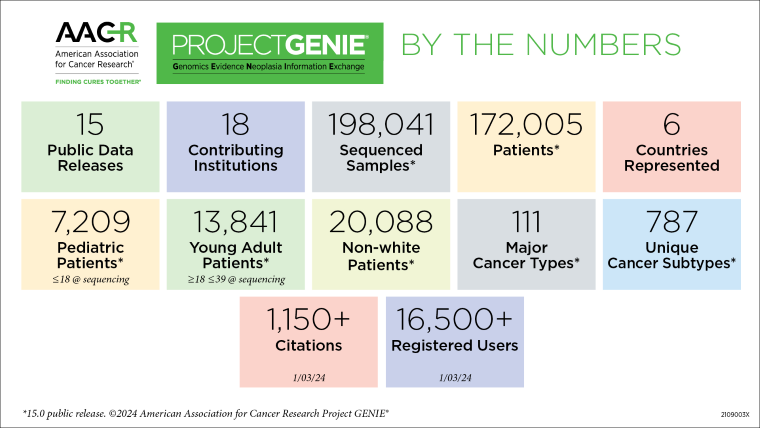
Launched in 2015, the AACR Project GENIE data registry offers cancer researchers data derived from diverse patient populations and cancer types harmonized from 22 participating institutions. With the release of Project GENIE 15.0 in January 2024 and the public release of two data sets (non-small cell lung cancer and colorectal cancer) from the Biopharma Collaborative (BPC) formed in 2019 with 10 biopharmaceutical companies, GENIE continues to expand its role in helping to drive clinical, translational, and computational cancer research. This was highlighted during two sessions at the AACR Annual Meeting 2024 that included presentations from several researchers who have utilized AACR Project GENIE.
One session was a minisymposium chaired by AACR Project GENIE Chairperson Philippe Bedard, MD, from UHN-Toronto General Hospital, with six presenters who shared how they used the database to advance cancer research. The other was a dedicated methods workshop chaired by Katherine Panageas, DrPH, of Memorial Sloan Kettering Cancer Center, that highlighted the methodological advancements and tools that AACR Project Genie now offers.
Both sessions helped put a new perspective on questions that have long plagued researchers.
How Do Some Patients’ Cancers Develop Resistance to KRAS Inhibitors?

Among the highlights from the minisymposium was research presented by Saikat Chowdhury, PhD, of the University of Texas MD Anderson Cancer Center, and his team on how activating PIK3CA mutations and the hedgehog signaling pathway could potentially confer resistance to KRAS inhibitors in colorectal cancer and lung adenocarcinoma (LUAD). They leveraged GENIE data alongside other sources to identify distinct co-mutation networks of KRAS in cohorts with these cancer types. They discovered that in colorectal cancer cohorts, KRAS co-mutated with PIK3CA, SMAD2, and FBXW7 genes while in the LUAD cohorts, KRAS co-mutated with STK11 and KEAP1.
They corroborated their findings with the data from The Cancer Genome Atlas (TCGA) as well as cell-based validation experiments. Their finding implies that other mutational pathways, apart from KRAS, may cause differential responses to targeted therapy against KRAS in colorectal cancer and LUAD patients, thereby conferring resistance to KRAS-targeted therapies.
Can Variants of Unknown Significance Be Reclassified?

Thinh Tran, of Memorial Sloan Kettering Cancer Center, and her team presented work on leveraging AlphaMisSense—a computational Variant Effect Predictor (VEP) tool—to reclassify variants of unknown significance (VUS) into pathogenic and nonpathogenic/benign categories. This was corroborated with real-world data obtained from the GENIE database to reclassify pathogenic variants associated with worse outcomes in non-small cell lung cancer.
This tool is particularly important considering 80% of variants identified by other known platforms, such as OncoKB, are classified as VUS, so the ability to reclassify these variants will play a major role in identifying the best treatment for patients. This shows that real-world data, such as from the GENIE database, can be used as benchmarks to evaluate the predictions of ML models regarding the effect of mutations and ultimately drive effective diagnosis and timely treatment of cancer patients.
How Does Lung Cancer Differ in Smokers and Nonsmokers?

Krishna Dasari, of the Yale School of Medicine, and team used GENIE data to explore the adaptive landscape of LUAD genomic mutations, shedding light on crucial differences between adenocarcinoma in smokers versus nonsmokers. Smoking leads to chronic inflammation, stress, altered immunity, and airway narrowing in the lungs. This results in both physiological and genetic alterations in the lung and is a major risk for lung cancer. LUAD, however, also develops in nonsmokers, so understanding the differences in driver mutations for smokers and nonsmokers is crucial to developing the best targeted therapy for their management and treatment.
In their study, mutational selection was observed to be different in smokers compared to nonsmokers, with KRAS, KEAP1, and STK11 mutations being stronger drivers of LUAD in smokers while EGFR, PIK3CA, and SMAD4 mutations were stronger drivers of LUAD in nonsmokers. The different adaptive landscape of LUAD between these two patient groups highlights the need to consider not just gene mutations in isolation, but the entire genetic map of the tumor, to develop optimal treatment regimens and inform precision oncology.
What Other Queries Can AACR Project GENIE Help Solve?
From the minisymposium, other presenters also displayed the value of GENIE data to their research, including:
- Ino de Bruijn, of Memorial Sloan Kettering Cancer Center, provided a comprehensive overview of the cBioPortal cancer genomics platform, which enables access to GENIE cohort, demonstrating its utility in helping to drive biological discovery and clinical applications.
- Sara Horie, of Keio University in Japan, presented findings from a pan-cancer study comparing mutational signatures of tumors from Asian patients from the Center for Genomics and Cancer Therapeutics (C-CAT) Japan database with those from white patients from the AACR GENIE and TCGA databases. She and her team found mutational frequencies in certain genes differed by race, which may influence disease biology and treatment outcomes.
- Tomohiro Tamura, of Keio University in Japan, and colleagues used GENIE data to conduct a molecular characterization of high-grade serous carcinoma (HGSC, the most common type of ovarian cancer). Their work identified a unique molecular signature specific to HGSC that may hold promise in identifying more precise targets for managing refractory HGSC in patients.
Meanwhile, the speakers in the workshop revealed the versatility and utility of GENIE data in other ways, including:
- Alex Paynter, a biostatistician from Sage Bionetworks supporting the GENIE project, highlighted the utility of real-world data sets and their outcomes on clinical research and U.S. Food and Drug Administration approvals, such as how GENIE data was included as part of the regulatory approval of sotorasib for KRAS-G12C mutant non-small cell lung cancer.
- Samantha Brown and Panageas, both from Memorial Sloan Kettering, presented an overview of “genieBPC,” which was developed as a resource to assist GENIE BPC data users in creating analytical data sets and associated summary statistics of patient outcomes. Their work was published in Bioinformatics in 2022.
- Bin Zhu, PhD, of the National Cancer Institute, presented work centered on pan-cancer mutational analyses using the Signature Analyzer for Targeted Sequencing (SATs). Zhu and colleagues demonstrated the versatility and utility of SATs in deciphering mutational landscapes across diverse cancer types and provided valuable insights into the underlying biology of cancer and its implications for patient management.
As we look to the future, the continued expansion and refinement of the AACR Project GENIE registry promises to pave the way for more personalized and effective treatments for cancer patients everywhere.
Jumi Popoola, PhD, is Scientific Program Manager with the AACR Project GENIE Coordinating Center.