Prostate Cancer Disparities: Race-related Biological Differences
Non-Hispanic black men in the United States are much more likely to develop prostate cancer and to die from the disease than their non-Hispanic white counterparts. Many factors contribute to this striking disparity, including access to and use of health care, social and economic status, and biology. As discussed by Steven R. Patierno, PhD, and colleagues in a recent perspective article in the AACR journal Clinical Cancer Research, alternative RNA splicing is one biological factor contributing to prostate cancer disparities.
Patierno will discuss additional insight into the role of alternative RNA splicing in race-related differences in prostate cancer when he gives his presentation in the Clinical and Translational Research in Diverse Populations plenary session being held on Monday, April 1, during the AACR Annual Meeting 2019.
Prostate cancer disparities
Non-Hispanic black men disproportionately shoulder the burden of prostate cancer in the United States. One in seven non-Hispanic black men are expected to develop prostate cancer during their lifetimes, compared with one in nine non-Hispanic white men, according to a recent report. Even more striking is the fact that non-Hispanic black men are almost twice as likely to die from the disease as their non-Hispanic white counterparts; one in 25 non-Hispanic black men compared with one in 45 non-Hispanic white men will die from prostate cancer.
Understanding the reasons for prostate cancer disparities is essential if we are to develop and implement strategies that will eliminate the disparities. However, as Patierno, who is deputy director of the Duke Cancer Institute, wrote in the abstract for his presentation, “Cancer disparities are the result of a complex interplay among social, structural (health system), lifestyle, and biological determinants of health.”
One area of intensive research investigation into biological determinants of prostate cancer disparities is identifying differences in genetic mutations between prostate cancer patients of different ancestry. Another source of biological heterogeneity is alternative RNA splicing, and this is the research focus of Patierno’s presentation, which is titled, “Spliceomics: Alternative RNA splicing as a source of ancestry-related molecular targets in precision oncology and cancer disparities.”
Alternative RNA splicing 101
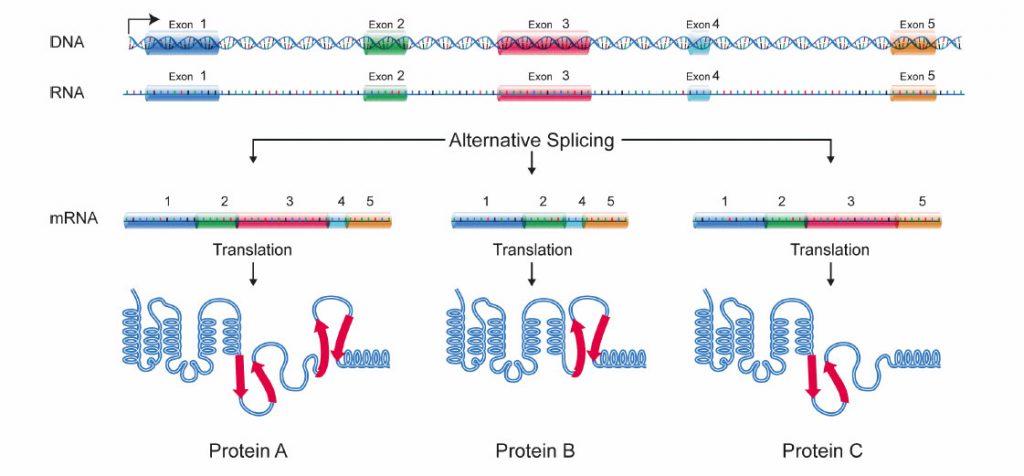
It is estimated that humans have between 20,000 and 25,000 genes. Genes contain the information a cell needs for producing the proteins the cell requires to function. Genes, which are made of DNA, are decoded into proteins through an intermediate product known as RNA.
Not all the DNA bases in a gene code for proteins. Genes have several protein-coding regions called exons separated by noncoding regions called introns. In the first step of the decoding process, the entire gene, including the exons and introns, is transcribed from DNA to RNA. This RNA is then processed to form the final protein-coding messenger RNA (mRNA). One of the processing steps, referred to as splicing, involves the exclusion of the introns. During splicing, some of the exons may also be excluded. Thus, several alternative mRNA molecules can be generated from a single gene.
Researchers estimate that more than 90 percent of human genes undergo alternative RNA splicing. This process provides a way for cells to have a more diverse array of proteins than would be possible if each gene could encode only one protein. In fact, it has been estimated that the complexity resulting from alternative splicing provides an explanation for how humans can have more than 250,000 proteins despite having just 20,000 to 25,000 genes.
Alternative RNA splicing, cancer, and prostate cancer disparities
In the Clinical Cancer Research perspective article, Patierno and colleagues discuss how alternative RNA splicing is emerging as an important source of heterogeneity in cancer, including prostate cancer, and how alternative RNA splicing can influence the aggressiveness and therapeutic responsiveness of a tumor. They also highlight that there are race-related differences in alternative splicing and suggest that these differences provide potential new therapeutic targets for precision medicines that could help mitigate cancer disparities among racial groups.
Regarding prostate cancer, the authors emphasize one prior study investigating RNA splice variants in prostate cancers obtained from non-Hispanic black and non-Hispanic white men. In that study, numerous race-specific RNA splicing variants were identified and several of the RNA splicing variants enriched in prostate cancers from non-Hispanic black men were found to drive race-related prostate cancer aggressiveness and influence tumor response to relevant therapeutics. For example, an RNA splice variant of PI3Kδ enriched in non-Hispanic black prostate cancers increased proliferation and invasive capacity in vitro compared with the non-Hispanic white PI3Kδ RNA splice variant, and it conferred resistance to a PI3Kδ-targeted therapeutic in a mouse model of prostate cancer.
In the perspective article, Patierno and colleagues go on to discuss the potential for harnessing our growing understanding of the role of alternative RNA splicing in cancer for therapeutic purposes. The first splicing-targeted therapeutic was approved by the U.S. Food and Drug Administration in December 2016, for treating patients with spinal muscular atrophy, a rare disease caused by mutations in the SMN1 gene that impair SMN protein production. The therapeutic, which is called nusinersen (Spinraza), is an antisense oligonucleotide that increases production of full-length SMN protein by increasing the proportion of SMN2 mRNA transcripts that include exon 7. Despite the proof-of-principle that this approval provides, Patierno and colleagues point out that relatively limited effort has been put into investigating therapeutic approaches to manipulating alternative splicing in cancer drug development.
Patierno will talk about new work being conducted in his laboratory to therapeutically manipulate alternative RNA splicing, correct aberrant RNA splicing, and produce novel RNA splice variants in models of prostate cancer and other types of cancer, as part of his presentation at the AACR Annual Meeting 2019. His hope is that these novel therapeutic approaches can drive race-stratified clinical trials and improve outcomes for patients, thereby reducing prostate cancer disparities and disparities in other types of cancer.