SABCS 2023: How Do Triple-negative Breast Cancers Evolve?
The key principles of cancer evolution — encompassing tumor heterogeneity, clonal evolution, chromosomal instability, and maintenance of truncal events in metastases — were described in an elegant paper by W. Fraser Symmans, MD in 1995.
Technological advances in the intervening decades have allowed researchers to gain better insight into cancer evolution.
“Simply put, the tools we’ve got nowadays allow us to elucidate this process in high definition,” began Charles Swanton, MD, PhD, FAACR, deputy clinical director of the Francis Crick Institute, who delivered a plenary lecture titled, “Breast cancer evolution, immune evasion, and metastasis driven by chromosomal instability” at the 2023 San Antonio Breast Cancer Symposium, held December 5-9.

Swanton is an architect and leader of the translational research study called Tracking Cancer Evolution Through Therapy (Rx) (TRACERx). Established in 2014, this prospective study aims to understand the spatiotemporal evolutionary trajectories of cancer, with the goal of furthering precision medicine. The investigators of the consortium have published several pivotal longitudinal studies outlining the factors behind lung and renal cancer progression.
Swanton and colleagues applied their extensive discoveries about lung cancer evolution from the lung TRACERx studies to ask important questions about the evolution of breast cancer in a large, multi-institutional study, the breast TRACERx. They queried early-stage triple-negative breast cancer (TNBC) tumors from 175 patients who received neoadjuvant treatment.
Devoting his presentation to the role chromosomal instability plays in breast cancer evolution, Swanton elaborated on the numerical aspects of instability—such as gains, losses, and doubling of chromosome number—and the structural aspects of instability—such as deletions, amplifications, inversions, and translocations. The discussion focused on the consequential role of truncal mutations—mutations that arise in the “trunk” of the evolutionary tree and play an important role in the tumor’s evolutionary trajectory, and subclonal mutations—acquired mutations in the “branches,” present in a subset of the tumor cells.
Several crucial observations were made from the pre-neoadjuvant treatment samples of TNBC. First, about one-fourth of the samples had at least one subclonal driver mutation, a phenomenon associated with poorer outcomes in the lung TRACERx study. Second, the most common subclonal driver mutations were in the genes PTEN, PIK3CA, RB1, and NF1. Third, mutations in the tumor suppressor gene TP53 were mostly truncal events, cementing their role as major drivers of chromosomal instability. Fourth, there were several routes to generating cancer diversity. A lack of clear correlation between mutational heterogeneity and chromosomal instability—meaning tumors with varying levels of mutational heterogeneity and chromosomal instability—provides tumors with the choice between the two to facilitate natural selection.
Drivers of genomic instability
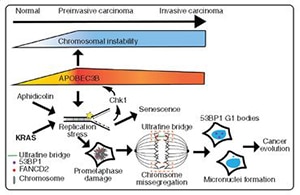
“Interestingly, and worryingly, there is extensive whole-genome doubling in TNBC—about 70%,” Swanton said, with about 2% of cancers having undergone two whole-genome doubling events, a clonal and a subclonal event. Whole-genome doubling is a major driver of chromosomal instability, Swanton said.
But why are whole-genome doubling events important for cancer cells?
Chromosomal instability leads to loss of heterozygosity (LOH), where a copy of an essential gene, such as TP53, is lost. This means that a mutation in the single remaining copy of the gene will inevitably lead to cell death. Cancer cells utilize whole-genome doubling as a clever workaround to this problem because they can weather a deleterious mutation in one of the two copies of the gene, thus enabling the cancer cell to prevail.
In light of this observation, Swanton and colleagues hypothesized that the frequency of whole-genome doubling will correlate with the proportion of genome loss to LOH. “And that is indeed the case,” Swanton said, noting that they observed a loss of nearly 30% of the TNBC genome, putting TNBC among the top cancer types with high LOH matched with a high frequency of genome doubling.
Similar to an observation made in lung cancer evolution, which Swanton and colleagues describe in their paper in Cancer Discovery, mutations in the gene-editing enzyme APOBEC3B were found to drive mutational diversity and chromosomal instability in TNBC. In their paper, TRACERx consortium members describe data from preclinical studies and patient samples that point to APOBEC3B induction and associated chromosomal diversity in preinvasive disease, thus setting the stage for cancer evolution.
Another feature of TNBC evolution is extrachromosomal DNA (ecDNA)-driven chromosomal instability. ecDNA, small, circular DNA structures residing outside the chromosomes, can be a major source of gene amplification. Owing to their lack of centromeres, they play an important role in a non-Darwinian mode of evolution through random segregation during mitosis, generating high gene copy number, ultimately leading to tumor heterogeneity.
ecDNAs are more frequent in TNBC than in lung cancer, Swanton’s team found, with a third of TNBCs harboring them.
What are the consequences of genomic instability?
In addition to ecDNAs being associated with poor survival for patients with TNBC, Swanton and colleagues made a novel observation: While about half of ecDNAs encode for oncogenes, the other half encode for immunomodulatory genes, promoting immune suppression. Genomic instability also drives loss of tumor suppressor genes early in the process of tumor evolution, which is followed by whole-genome doubling and amplification of oncogenes.
Other outcomes of genomic instability are HLA loss and immune evasion. Chromosomal instability, and ensuing HLA LOH, impairs HLA class I-mediated tumor antigen recognition and binding, which is essential for presenting the tumor antigen to T-cell receptors. Swanton also discussed novel data on the role alternative splicing of HLA molecules plays in impeding neoantigen presentation in estrogen receptor-positive breast cancer.
“As a result of this propagation of chromosomal instability … diversity ensues, natural selection acts,” concluded Swanton. “This results in increased proliferation and diversity of proliferation indices across TNBC subclones, parallel evolution of events that are manifested as selection in metastases, and immune evasion events that include HLA LOH and ecDNA-driven immune suppression.”



